Page 77 - 南京医科大学自然版
P. 77
第44卷第7期 王 皓,杜昱升,王 友,等. 基于光谱CT和影像组学特征的机器学习模型和列线图预测甲
2024年7月 状腺结节良恶性[J]. 南京医科大学学报(自然科学版),2024,44(7):958-965 ·961 ·
表1 训练集与验证集的基本信息
Table 1 Basic information of the training and validation sets
2
Information Training set(n=100) Validation set(n=43) Total(n=143) χ /t/Z P
Clusters[n(%)] 0.794 0.068
Benign 31(31) 15(35) 046(32)
Malignant 69(69) 28(65) 097(68)
Sex[n(%)] -0.104 0.747
Female 83(83) 34(79) 117(82)
Male 17(17) 09(21) 026(18)
Age(years,x ± s) 53.00 ± 13.73 49.14 ± 16.63 51.84 ± 14.71 -1.339 0.185
NIC(x ± s) 0.56 ± 0.19 0.57 ± 0.20 0.56 ± 0.19 0.383 0.705
λHu [M(P25,P75)] 2.83(2.09,3.64) 2.85(2.21,3.33) 2.85(2.12,3.58) -0.288 0.773
FT3(pmol/L,x ± s) 4.68 ± 0.69 4.64 ± 0.60 4.67 ± 0.66 -0.325 0.746
FT4[pmol/L,M(P25,P75)] 16.45(14.17,18.80) 15.15(14.00,19.30) 16.30(14.10,18.90) -0.326 0.745
TSH[mU/L,M(P25,P75)] 1.72(1.33,2.78) 1.53(1.21,2.64) 1.65(1.27,2.74) -0.564 0.573
A 36 33 16 6 3 B 6056544540363531302523201811 8 10 8 7 6 6 6 5 4 4 2 2 2 1
3
0.26
2
0.24
1 0.22
Coefficients -1 0 Mean⁃squared error 0.20
-2 0.18
-3 0.16
-4
0.14
-6 -5 -4 -3 -2 -6 -5 -4 -3 -2
ln(λ) ln(λ)
C D
Original_firstorder_Minimum
3
***
Original_firstorder_10Percentile
2
Original_glszm_LowGrayLevelZoneEmphasis 1
Radscore 0
Log.sigma.3.0.mm.3D_firstorder_90Percentile
-1
Original_ngtdm_Busyness
-2
Log.sigma.4.0.mm.3D_glcm_Id
-3
-0.2 0 0.2 0.4
Benign Malignant
Weight
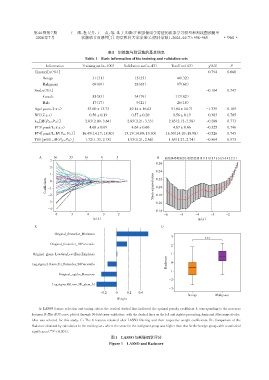
A:LASSO feature selection and tuning,where the vertical dashed line indicated the optimal penalty coefficient λ corresponding to the non⁃zero
features. B:The AUC curve plotted through 10⁃fold cross⁃validation,with the dashed lines on the left and right representing λmin and λ1se respectively,
λ1se was selected for this study. C:The 6 features retained after LASSO filtering and their respective weight coefficients. D :Comparison of the
Radscore obtained by calculation in the training set,where the score for the malignant group was higher than that for the benign group,with a statistical
***
significance( P < 0.001).
图1 LASSO与影像组学评分
Figure 1 LASSO and Radscore