Page 56 - 南京医科大学学报自然科学版
P. 56
第44卷第3期
·346 · 南 京 医 科 大 学 学 报 2024年3月
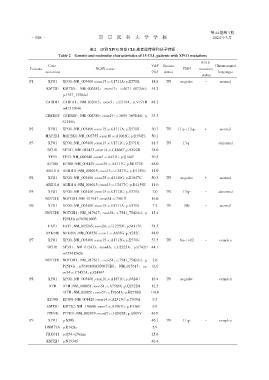
表2 15例XPO1突变CLL患者遗传学和分子特征
Table 2 Genetic and molecular characteristics of 15 CLL patients with XPO1 mutations
IGHV
Gene VAF Disease Chromosomal
Patients HGSV name FISH mutation
mutations (%) status karyotype
status
P1 XPO1 XPO1:NM_003400:exon15:c.G1711A:p.E571K 14.8 TN negative - normal
KMT2D KMT2D: NM_003482: exon13: c.4071_4073del: 51.2
p.1357_1358del
CARD11 CARD11:NM_032415:exon5:c.G511A:p.V171M 48.2
rs41319046
CREBBP CREBBP:NM_004380:exon19:c.3685_3698del:p. 33.3
S1229fs
P2 XPO1 XPO1:NM_003400:exon15:c.G1711A:p.E571K 30.7 TN 11q-;13q- + normal
MAP2K1 MAP2K1:NM_002755:exon10:c.A1062C:p.Q354H. 50.1
P3 XPO1 XPO1:NM_003400:exon15:c.G1711C:p.E571Q 21.5 TN 13q- - abnormal
SF3B1 SF3B1:NM_012433:exon14:c.G1866T:p.E622D 38.0
TP53 TP53:NM_000546:exon5:c.A431C:p.Q144P 30.5
EP300 EP300:NM_001429:exon29:c.A4717G:p.M1573V 48.0
ARID1A ARID1A:NM_006015:exon13:c.C3477G:p.D1159E 11.0
P4 XPO1 XPO1:NM_003400:exon25:c.A3140G:p.E1047G 50.3 TN negative + normal
ARID1A ARID1A:NM_006015:exon13:c.C3477G:p.D1159E 11.0
P5 XPO1 XPO1:NM_003400:exon15:c.G1712G:p.E571K 7.0 TN 13q- - abnormal
NOTCH1 NOTCH1:NM_017617:exon34:c.7501T 16.0
P6 XPO1 XPO1:NM_003400:exon15:c.G1711A:p.E571K 7.2 TN ND - normal
NOTCH1 NOTCH1:NM_017617:exon34:c.7541_7542del:p. 15.4
P2514fs rs763016003
FAT1 FAT1:NM_005245:exon24:c.C12350T:p.S4117L 51.5
NFKBIE NFKBIE:NM_004556:exon1:c.A635G:p.Y212C 48.0
P7 XPO1 XPO1:NM_003400:exon15:c.A1712G:p.E571G 37.5 TN 6q-;+12 - complex
SF3B1 SF3B1:NM_012433:exon16:c.G2225A:p.G742D 44.1
rs755415626
NOTCH1 NOTCH1:NM_017617:exon34:c.7541_7542del:p. 2.6
P2514fs rs763016003/NOTCH1:NM_017617:ex⁃ /2.1
on34:c.C7457A:p.S2486*
P8 XPO1 XPO1:NM_003400:exon16:c.A1871G:p.D624G 16.4 TN negative - complex
ATM ATM:NM_000051:exon51:c.A7566C:p.Q2522H 12.5
/ATM:NM_000051:exon57:c.T8364A:p.H2788Q /14.0
EP300 EP300:NM_001429:exon14:c.A2515G:p.T839A 9.3
KMT2C KMT2C:NM_170606:exon7:c.A1007G:p.E336G 6.9
PTPRD PTPRD:NM_002839:exon27:c.G2426T:p.G809V 46.9
P9 XPO1 p.N30S 46.1 TN 11q- - complex
DNMT3A p.R742fs 5.9
FBXO11 p.Q54-q56dup 25.6
KMT2D p.N1934S 46.4