Page 36 - 南京医科大学自然版
P. 36
第44卷第6期
·772 · 南 京 医 科 大 学 学 报 2024年6月
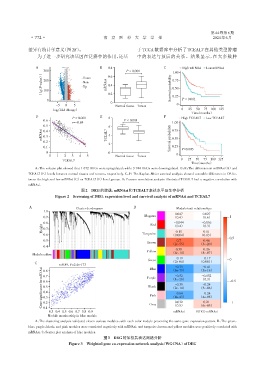
差异有统计学意义(图2F)。 于 TCGA 数据库中分析了 TCEAL7 在其他类型肿瘤
为了进一步研究该基因在泛癌中的作用,还基 中的表达与预后的关系。结果显示,在大多数肿
A B 0.8 C High mRNAsi Low mRNAsi
| 300 Down 0.6 P < 0.001 1.00
(P⁃value) 200 Non mRNAsi 0.4 Survival probability 0.75
Up
0.50
|lg 100 0.2 0.25
0 P < 0.001
0
-5 0 5 Normal tissue Tumor 0
log2 (fold change) 0 25 50 75 100 125
Time(months)
D E F
P < 0.001 8 High TCEAL7 Low TCEAL7
0.6 P < 0.001
r=-0.84 1.00
0.5 6 0.75
mRNAsi 0.4 TCEAL7 4 Survival probability 0.50
0.3
0.2
2
0.1 0.25 P=0.003
0 0 0
0 1 2 3 4 5 Normal tissue Tumor
0 25 50 75 100 125
TCEAL7
Time(months)
A:The volcano plot showed that 1 672 DEGs were upregulated,while 3 744 DEGs were downregulated. B,E:The difference in mRNAsi(B)and
TCEAL7(E)levels between normal tissues and tumors,respectively. C,F:The Kaplan⁃Meier survival analysis showed a notable difference in OS be⁃
tween the high and low mRNAsi(C)or TCEAL7(F)level groups. D:Pearson correlation analysis illustrated TCEAL7 had a negative correlation with
mRNAsi.
图2 DEG的筛选,mRNAsi和TCEAL7表达水平及生存分析
Figure 2 Screening of DEG,expression level and survival analysis of mRNAsi and TCEAL7
A B
Cluster dendrogram Module⁃trait relationships
1.0
0.047 0.027
Magenta
0.9 (0.4) (0.6) 1
-0.044
-0.056
0.8 Red (0.4) (0.3)
Height 0.7 0.15 0.11
0.6 Turquoise (0.004) (0.03)
0.5
0.5 0.7 0.46
Brown
(2e⁃55) (3e⁃20)
0.4
0.38 0.26
Yellow (6e⁃14) (8e⁃07)
Module colors
-0.19 -0.17
Green 0
C (2e⁃04) (0.001)
r=0.89,P=2.4e⁃173 Blue (6e⁃79) (5e⁃16)
-0.41
-0.79
Gene significance for mRNAsi 0.6 Purple (1e⁃26) (3e⁃06) -0.5
0.8
-0.52
-0.052
0.7
(0.3)
-0.24
-0.39
Black
0.5
(2e⁃14)
0.4
-0.24
-0.64
Pink
(4e⁃43)
(4e⁃06)
0.3
0.032
0.21
0.2
(6e⁃05)
0.1
0.3 0.4 0.5 0.6 0.7 0.8 0.9 Grey (0.5) EREG⁃mRNAsi -1
mRNAsi
Module membership in blue module
A:The clustering analysis validated eleven various modules,with each color module presenting the same gene expression pattern. B:The green,
blue,purple,black,and pink modules were correlated negatively with mRNAsi,and turquoise,brown and yellow modules were positively correlated with
mRNAsi. C:Scatter plot analysis of blue modules.
图3 DEG的加权共表达网络分析
Figure 3 Weighted gene co⁃expression network analysis(WGCNA)of DEG